Science and art collide: Biomedical engineer Luca Borro reveals how he visualizes incredible organic molecules with V-Ray for Rhino and Chaos Cloud Rendering.
We’ve seen V-Ray used for all sorts of incredible projects, from Hollywood blockbusters to clothing simulators, but biomedical engineering is a new one. These incredible glimpses into microscopic worlds are created by biomedical engineer Luca Borro, a huge fan of V-Ray’s Interactive Rendering in Rhino and the seamless workflow of Chaos Cloud Rendering.
So when we met Luca, we were eager to learn how he implements our software to make microscopic molecules visible to the human eye.
We find it exciting that our software continues to unite professionals from various industries, including biomedicine, and gives knowledge to those that seek it. We sat down with Luca to discuss merging the scientific and artistic, and his workflow in V-Ray for Rhino.

About Luca
Luca Borro is a Biomedical Engineer that currently works at Bambino Gesù Children’s Hospital in Rome. His job involves the 3D reconstruction and photorealistic rendering of macromolecules of the human organism. He enjoys listening to pop and rock music, and reading while surrounded by the rolling hills of Umbria.
What got you interested in biology?
I am passionate about everything related to medicine and human biology.
Even though I earned my first degree in architecture, my real passion has always been medicine. In fact, I obtained a second degree in biomedical engineering and I am currently pursuing a master's degree in biology of health and nutrition at the University of L'Aquila in Italy.
I am fascinated by the physiology of the human body, the physiological activities of our organs, and the incredible complexity of our biological mechanisms.
Do you have any artistic training? How important is creativity and having a strong visual perception in your work?
As I said I obtained a first degree in architecture in 2013 which helped to develop my artistic sensibilities. Although I do not have a particular interest in the construction of buildings or building artifacts, the degree in architecture has provided me with important knowledge that has been useful to me in the biomedical field. During my studies as an architect, I learned a lot about descriptive geometry and 3D technology, which are two fascinating and extremely useful subjects. I then developed the ability to connect rational thinking with artistic thinking and creativity, which is really useful in life in general.
At what point did you begin incorporating 3D into your work as a biomedical engineer?
Immediately after my first degree in architecture, I thought it would be useful to bring 3D into the biomedical field. In 2013, 3D in medicine and biology was not as developed as it is today. The possibility of seeing molecules, organs, and tissues in three dimensions fascinated me so I started investigating the software and hardware tools needed to transform, for example, a radiological image into a 3D model. Later I discovered the world of molecular biology where 3D has a role not only in communication but in real research.
In fact, the organic molecules of our organism are protein structures with a precise three-dimensional shape that can be obtained from scanning technologies that make use of x-rays or, more recently, from sophisticated artificial intelligence algorithms. Even a small error in the 3D form of a certain protein can cause a serious defect in human health. This is absolutely amazing.
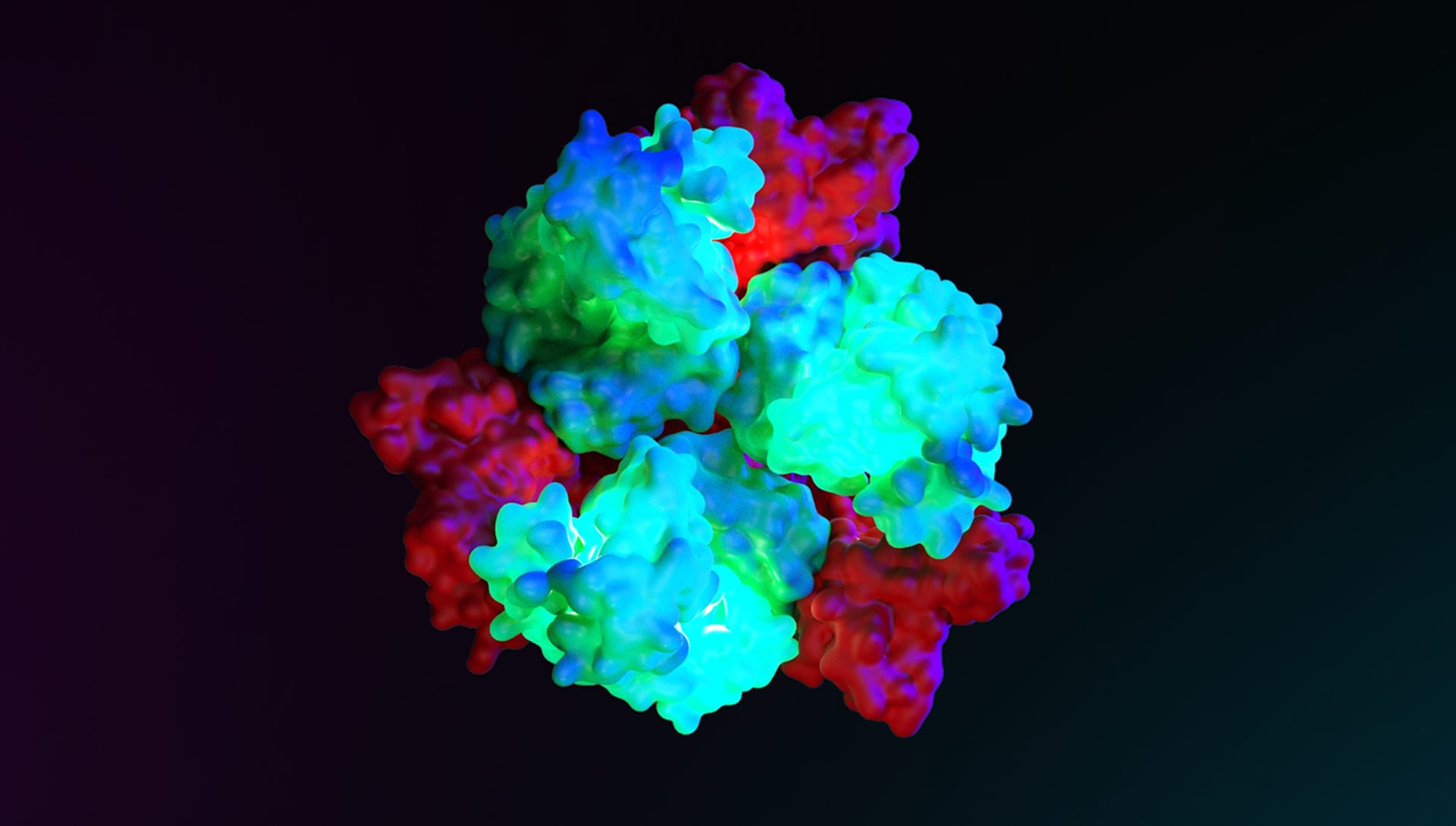
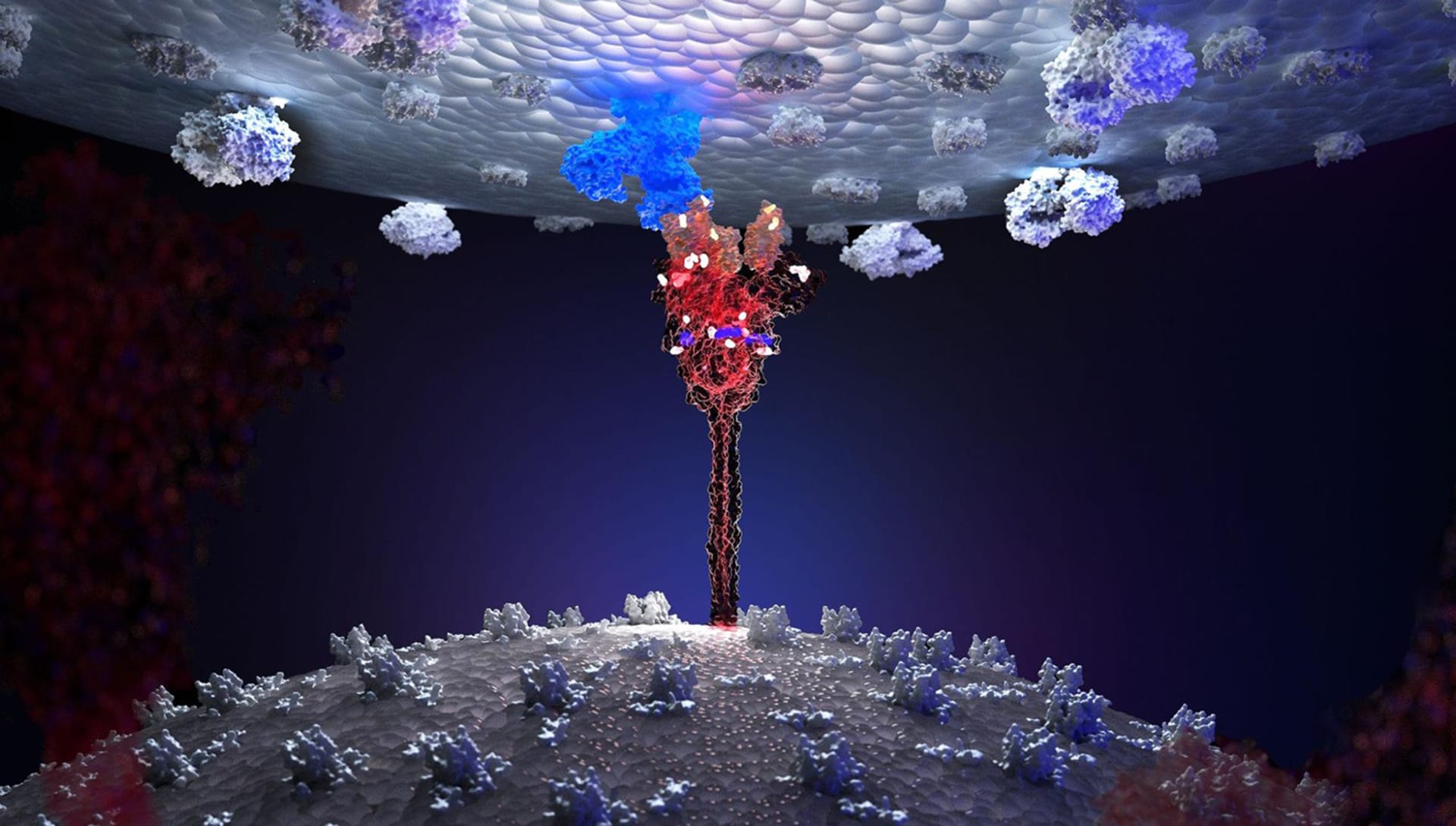
Your reconstruction of the Sars-CoV2 molecular structure was published on the cover of Elsevier’s Biomolecular Interactions scientific journal. That is an impressive achievement! How did that happen?
For this work, I was contacted by Dr. Antonella Di Pizio, a researcher at the Leibniz Institute for Food System Biology at the Technical University of Munich, who was very interested in the rendering techniques of the Sars-CoV-2 molecule. Antonella suggested that I describe in detail the realization of a photorealistic rendering of the molecule in order to publish this article in the scientific impacted journal Methods in Cell Biology. My work was even chosen for the cover of Volume 169 of the Biomolecular Interaction Part A series and this was a great satisfaction. The power of photorealistic rendering is truly incredible when applied to the representation of infinitely small human biological molecules.
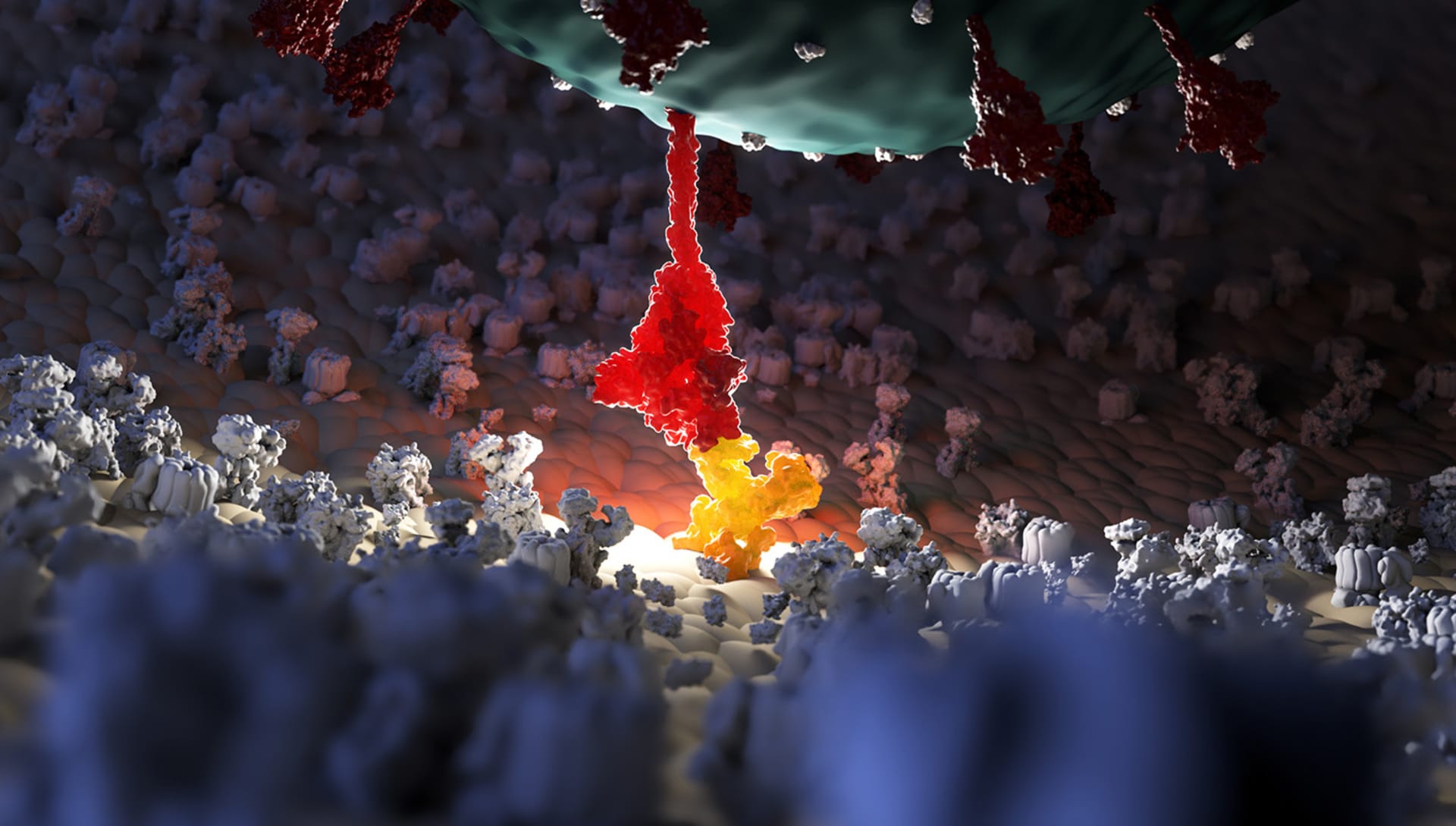
What level of research goes behind the creation of such a render? What part is left to artistic imagination?
This is a particularly important topic in scientific 3D representation.
When we want to represent in 3D a molecule of our organism (for example a protein) we have to deal with what the scientific literature makes available to this molecule.
Generally, by accessing databases that contain the protein structures in 3D, we are able to obtain the exact three-dimensional shape of the structure we intend to represent. However, it must be borne in mind that this structure is the result of the molecule scanning process through radiological imaging techniques such as X-ray crystallography.
These techniques may have a certain level of inaccuracy in the 3D reconstruction of the studied molecule so some parts of the molecule may not be well represented in 3D and could be missed in the final representation.
All the parts of the molecule that are well represented and those that are altered by "voids" or "holes" in the structure are thus explained on the 3D protein databases.
When we have to represent an “empty” part of the protein whose three-dimensional shape we do not know exactly, we cannot in any case rely on our artistic creativity but we must make visible the fact that that part of the molecule is not physically representable. We can do this by approximating the "empty" parts with elementary geometric shapes (for example with perfectly tubular shapes that make it clear that that part of the molecule is missing).
The artistic ideation is not allowed on the 3D structure of the protein which is already determined by science but it is instead possible on everything related to the rendering, the materials, the light design of the scene and, in general, the scenographic context in which you want to insert the protein.
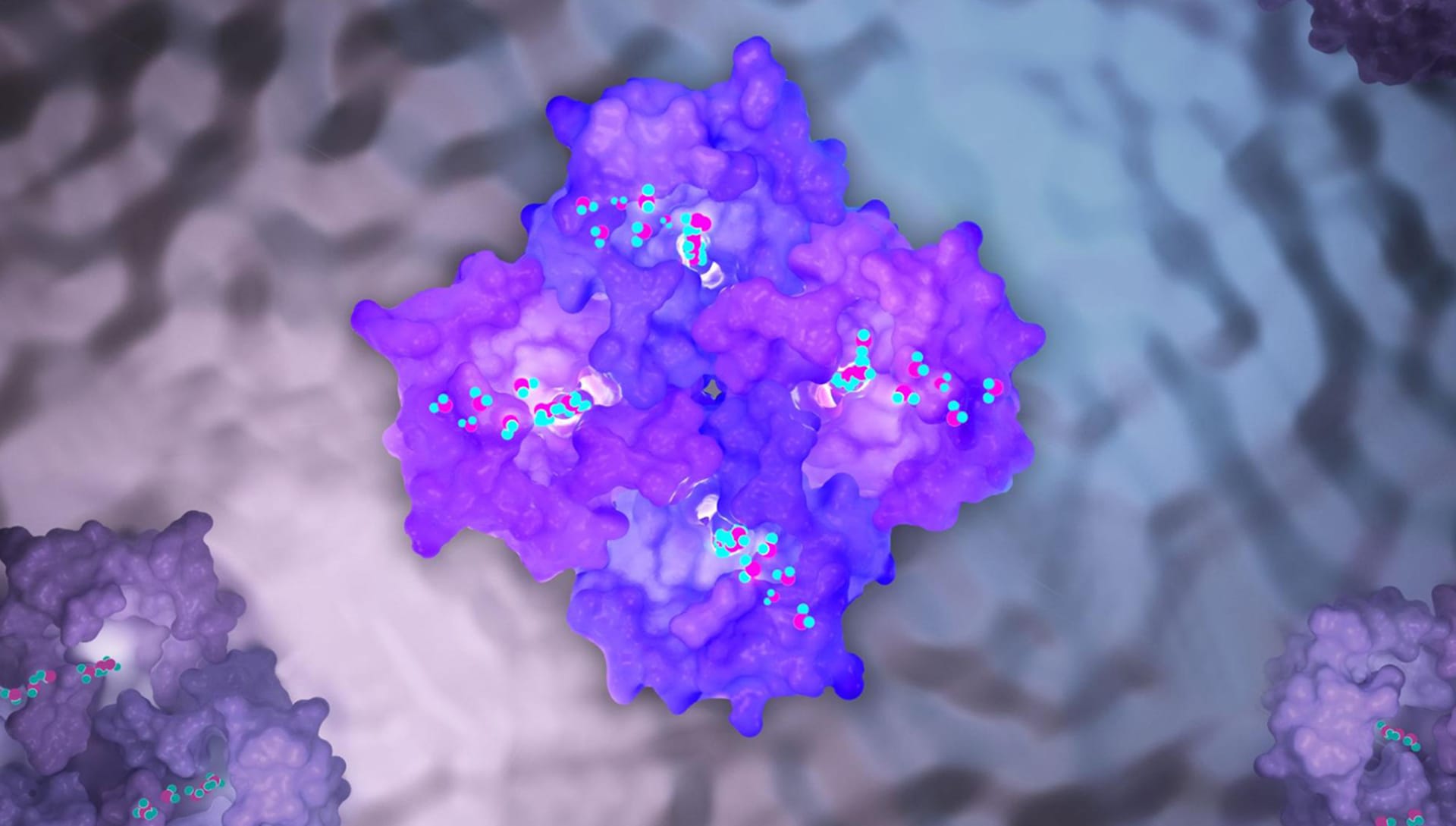
How did you model and texture the small intricate details in the cells?
The three-dimensional models of the complex molecules of our body are carefully described in .pdb files. We can compare them to common .stl or .obj files but these files contain complex information such as the coordinates of the atoms that make up the molecule, the amino acid sequences of the protein itself, and a number of other important bioinformatics tags.
Therefore, it is not the operator who manually models the protein in 3D but this is obtained from the scientific literature that deposits the 3D structures (and related files) of the proteins we know today on international platforms such as www.rcsb.org
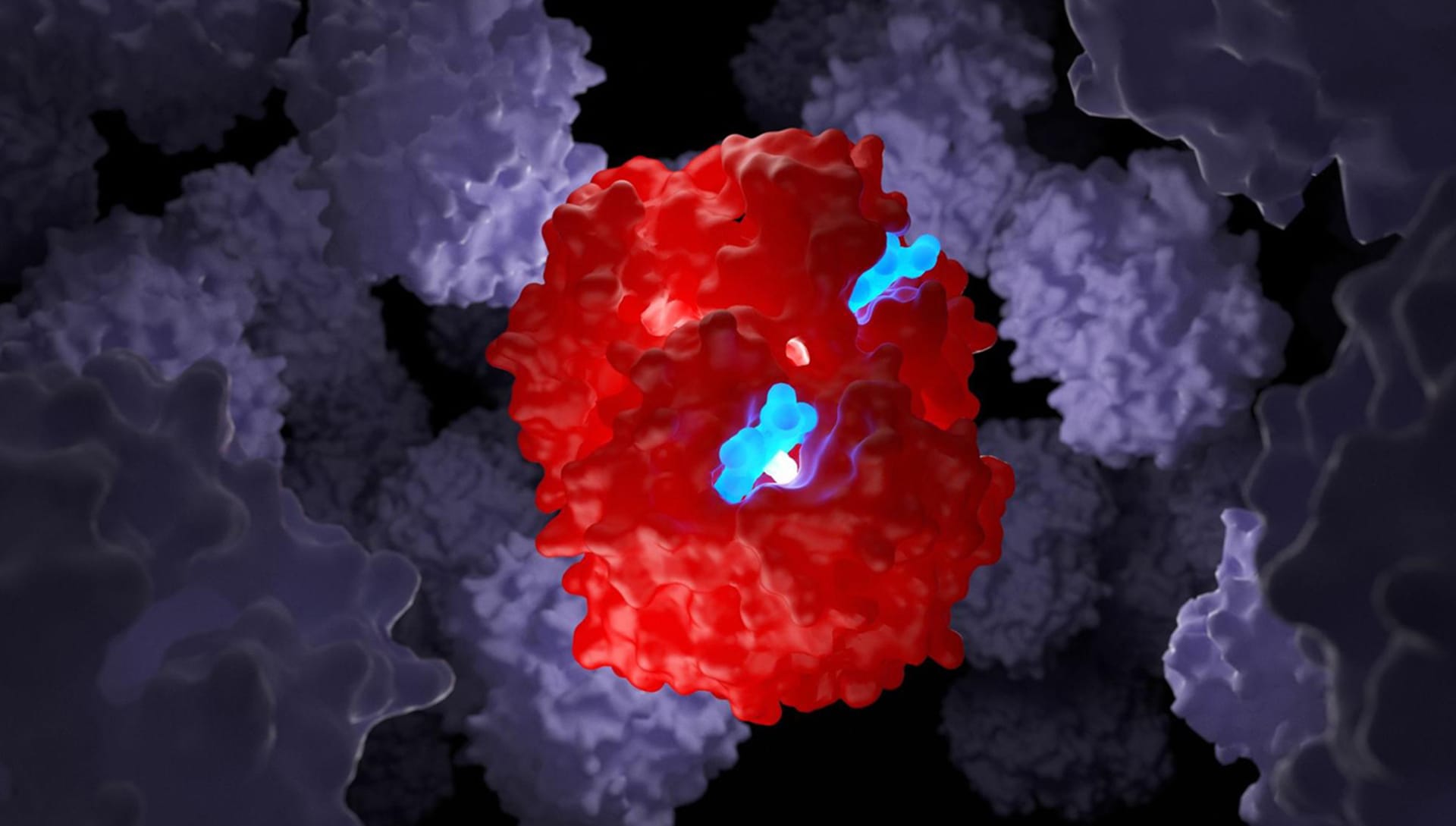
Can you share the process of creating the lighting?
Light is fundamental in our works. I am a big fan of light design, and if I had not followed the biological path it is very likely that I would have chosen to be a light designer for shows and theatrical performances.
I generally use two types of light elements in V-Ray: Plane Light and Sphere Light. Before "turning on" the lights, I collect my entire scene inside a large sphere to create complete darkness.Then I apply a total grey Override Materials to the entire scene and start the lighting design phase to test the final effect.
I need the lights to highlight some parts of the molecules that must seem "on" like lanterns. This helps me simulate the fact that these parts of the molecule are biologically "active" in a specific moment of the represented scene. For that, I use sphere lights within parts of the molecule to highlight and then I apply a Subsurface Scattering Material. I find this material incredible because it allows me to create photorealistic scenes exactly as I have them in mind.
Then I switch to the lights outside the molecule and tend to leave the main protein structure illuminated only in a few small specific points while the rest is usually almost completely dark. I use this theatrical effect in all my work.
The idea is to capture the viewer's eye only on the parts that I want to highlight. To do this, I bring a carefully calibrated sphere light close to the part of the molecule that I want to highlight. I don't use Spot-Light because I don't like the strength with which this element illuminates the molecule.
Then I move on to the background lighting. For this, I use Plane Light leaning against the walls of the sphere container. I generally use two or three sphere lights, two of which are placed sideways facing each other with two different complementary colors. I position the third higher to give a greater sense of depth.
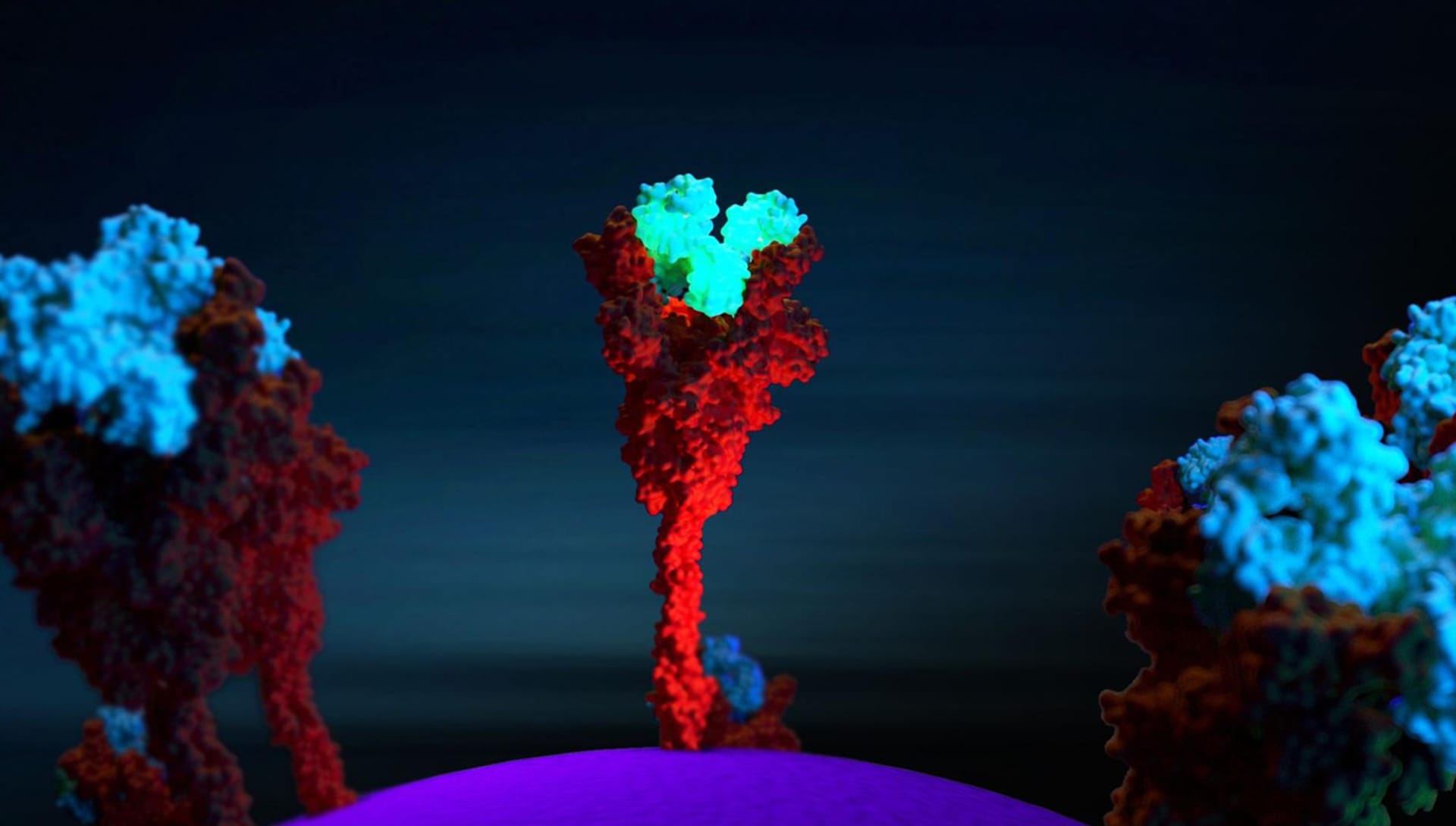
How do you approach composition in your work? Are there any mechanisms that you use?
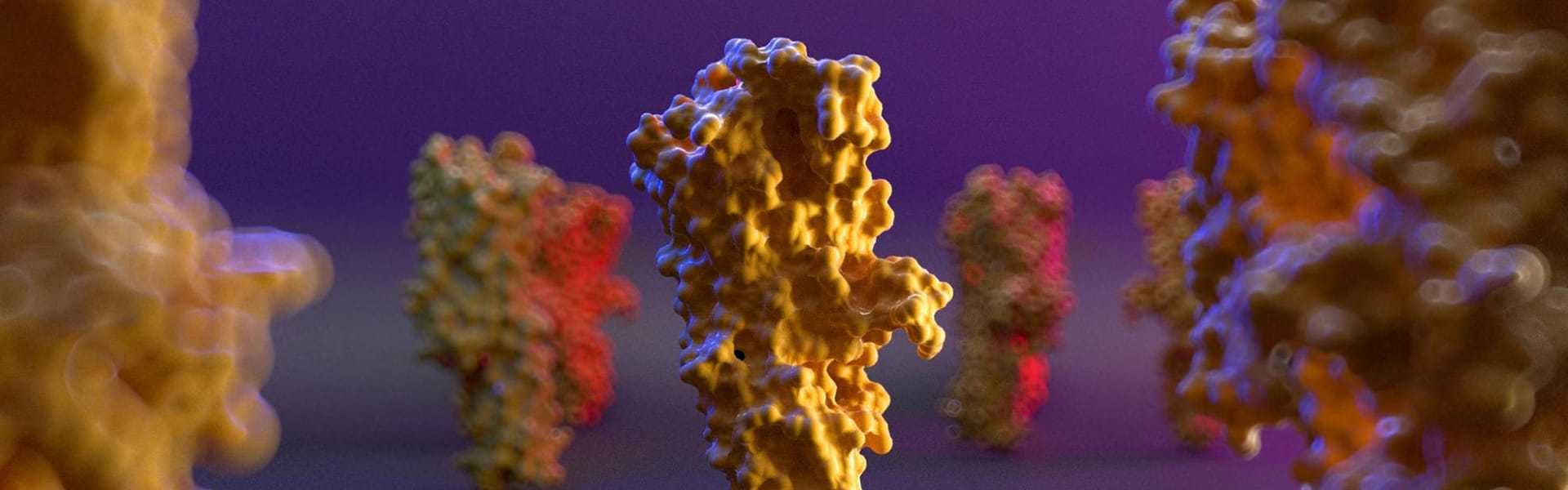
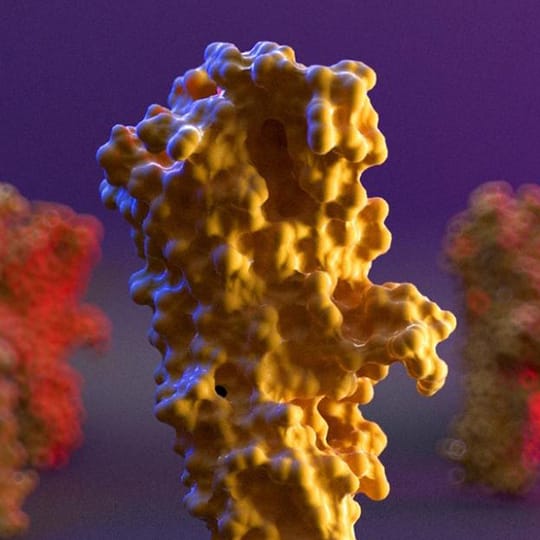
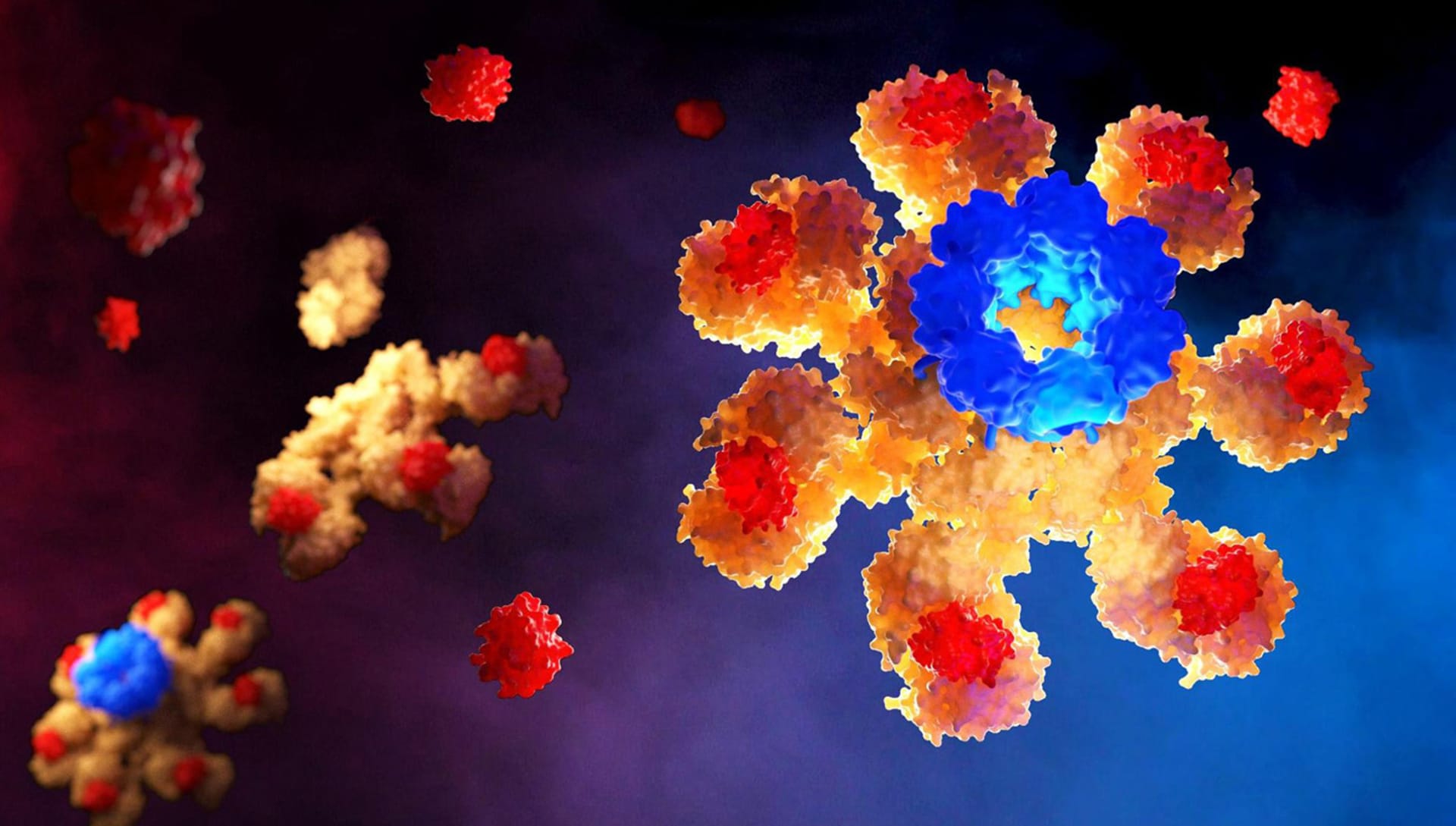
It really depends on the molecule I want to represent. In some molecules, the Rule of Thirds is fundamental, while for others I insert the molecule exactly in the center of the scene with the void around it. It is important to be able to represent the fact that in our organism the molecules that I represent are in continuous and very fast movement. What we see in the rendering is simply a single frame: a still image.
In reality, there would be continuous movement of the main molecule in the scene and of the other molecules around it due to the electrostatic interactions of the atoms that compose it. To do this I usually put in the foreground and in the background other molecules "in motion" which are blurred because I always set the focus with a point directly on the molecule in the area I want to keep in focus (Depth of Field - Fixed Point in Camera Settings).
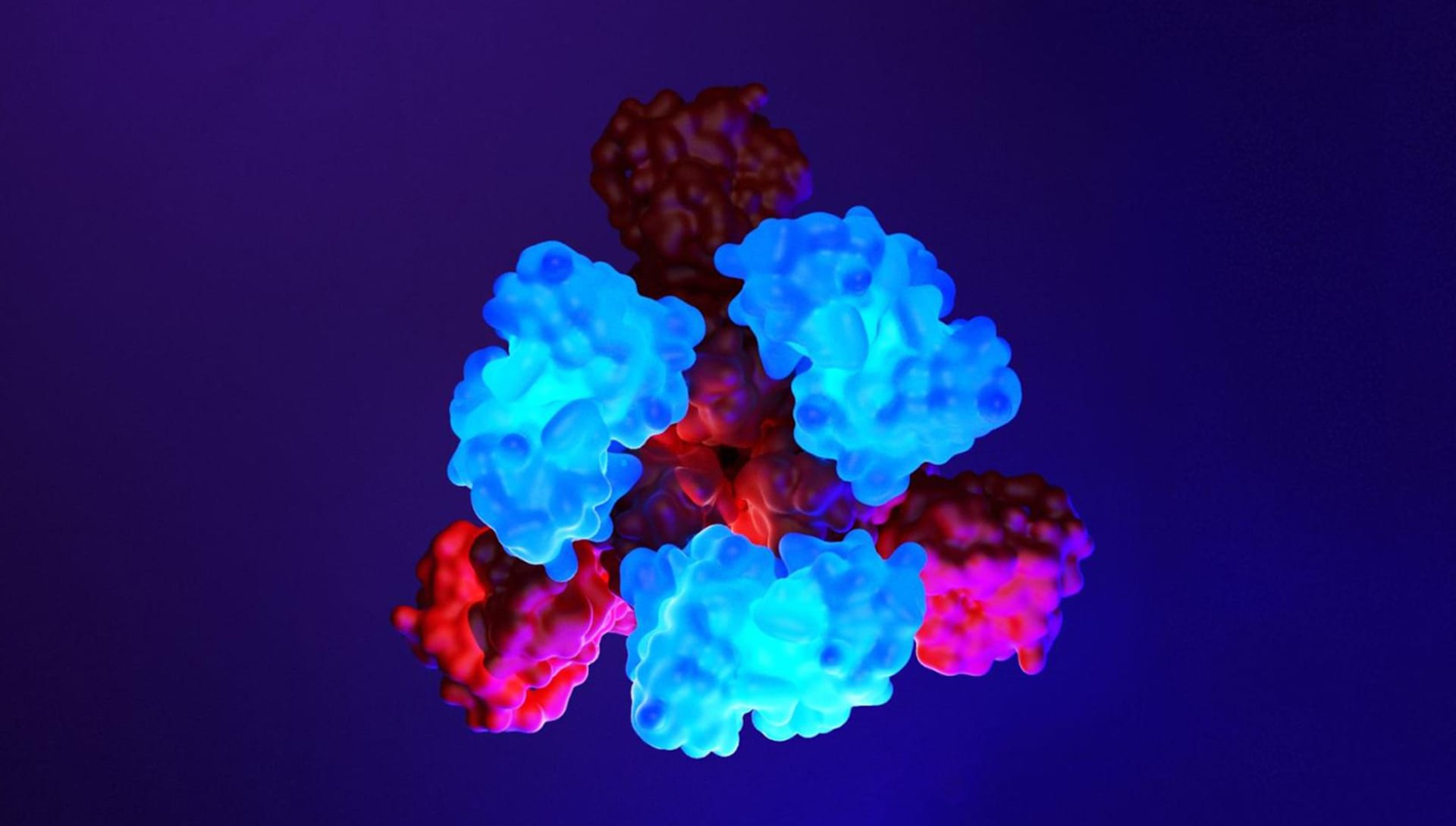
What is the most important factor to consider when rendering to achieve extreme biological accuracy?
You need to know how the molecule is made from the point of view of molecular biology. This means carefully studying the scientific literature that generally accompanies the 3D structure available on databases. By studying the scientific articles concerning the specific molecule it is possible to understand its structure, how it is made, its correct amino acid sequence, and therefore various important aspects can be understood such as where are the points in which that molecule binds with other molecules or with drugs. Biological accuracy in 3D representation can only be achieved if you avoid resorting to your own imagination in defining the 3D geometry of the protein and fully understand the biological structure of the molecule.
What is your favorite V-Ray for Rhino feature?
Interactive Rendering is essential for this type of work, but the possibility of rendering on the Cloud is fundamental. In biological laboratories, we do not have render farms capable of supporting complex high-resolution renders. For this reason, the ease and simplicity with which V-Ray for Rhino allows the control of the render in an interactive way and the subsequent rendering in the Cloud is really useful. The simple way in which it is possible to define the materials is also very useful, managing to immediately obtain good materials for the scene.
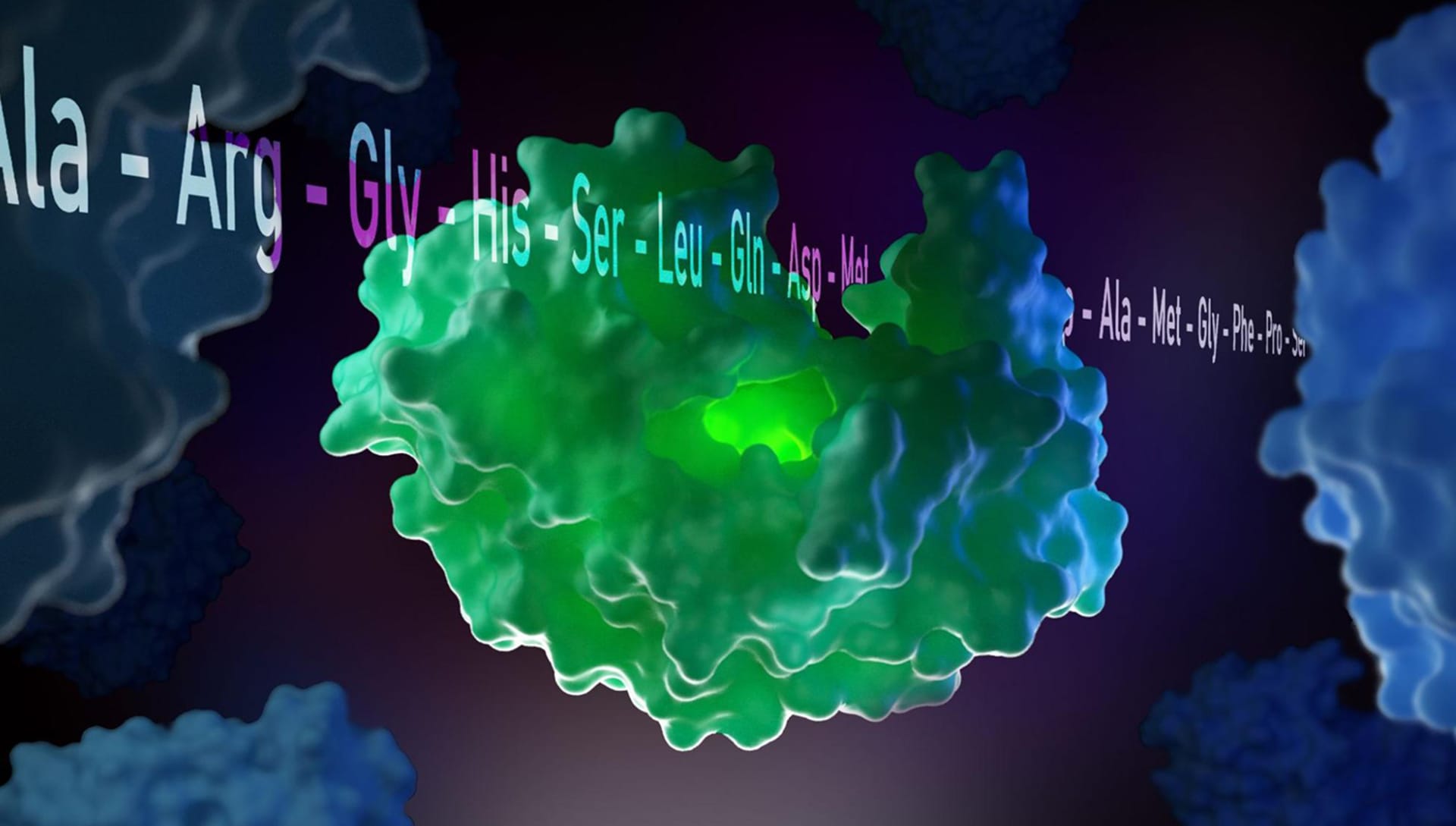
Similar to peer reviews in written journals, scientific illustrations and renderings must follow a strict validation process. Can you share it with us?
The validation process of an image can be performed by the operator himself who creates the scene or by other laboratories to which supervision is requested. In the case of the representation of the Sars-CoV-2 molecule, for example, I made use of the supervision of the Microbiology Laboratory of the Bambino Gesù Hospital in Rome where the molecule was carefully supervised.
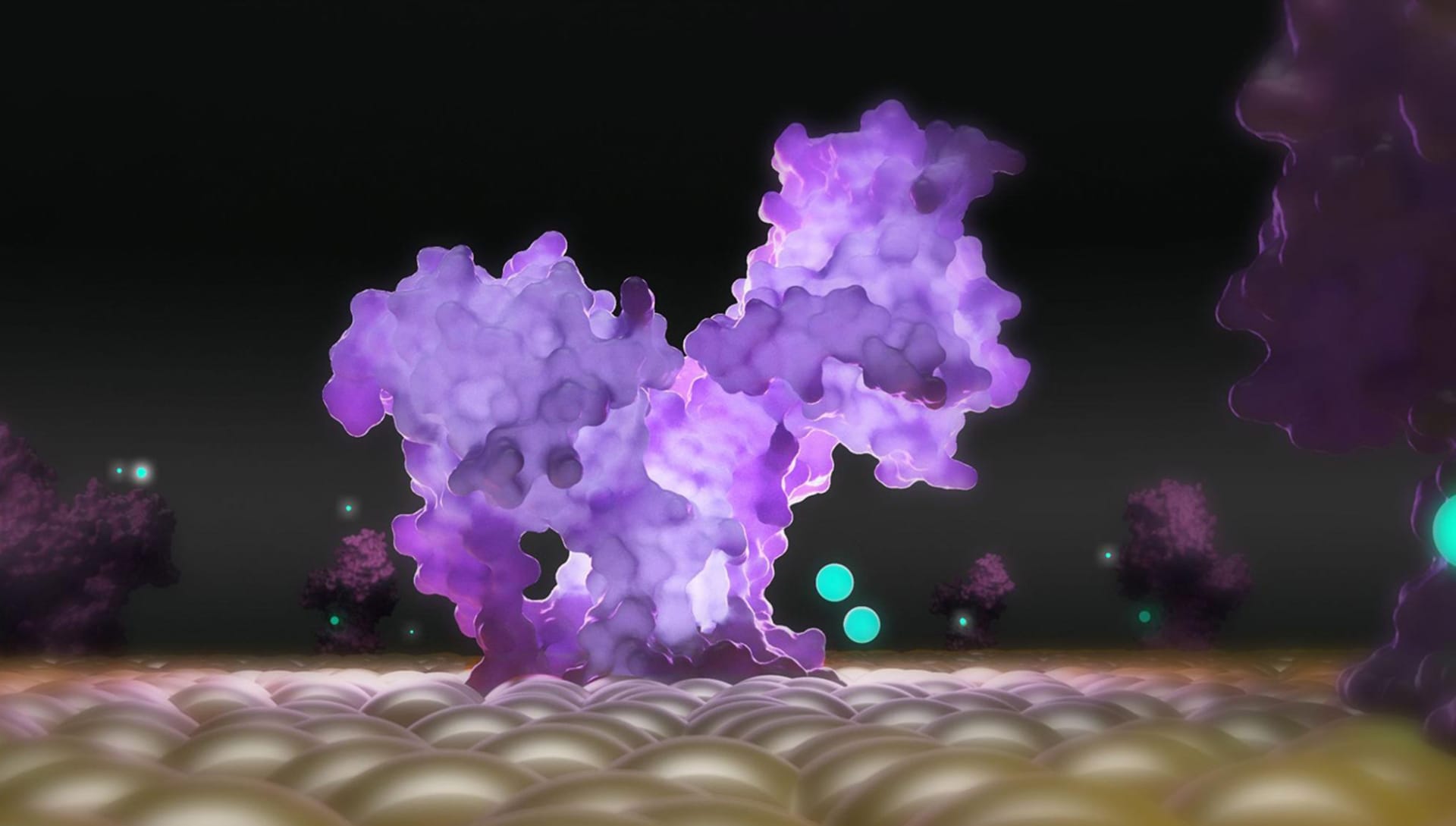
What projects do you have lined up next?
At the moment we are continuing the scientific dissemination collaboration of photorealistic molecular images with prestigious Italian magazine Focus. Subsequently, we will be able to organize webinars and workshops dedicated to this sector for all researchers and passionate operators in the field of biology and scientific visualization who are interested in learning more.